Originally from Scotland, Richard Benton received his PhD in 2003 from the University of Cambridge, and was an EMBO/Heley Hay Whitney post-doctoral fellow at The Rockefeller University, New York. He joined the Center for Integrative Genomics in September 2007 as Assistant Professor and promoted to Associate Professor in 2012 and Full Professor in 2018. His group’s research has been recognised by the Eppendorf & Science Prize for Neurobiology (2009), Friedrich Miescher Award (2012), AChemS Young Investigator Award for Research in Olfaction (2012), National Latsis Prize (2015) and EMBO Gold Medal (2016). His lab has been supported by the Swiss National Science Foundation, ERC Starting, Consolidator and Advanced Grants, EMBO and HFSP. He was elected EMBO member in 2019 and a Fellow of the Royal Society in 2021.
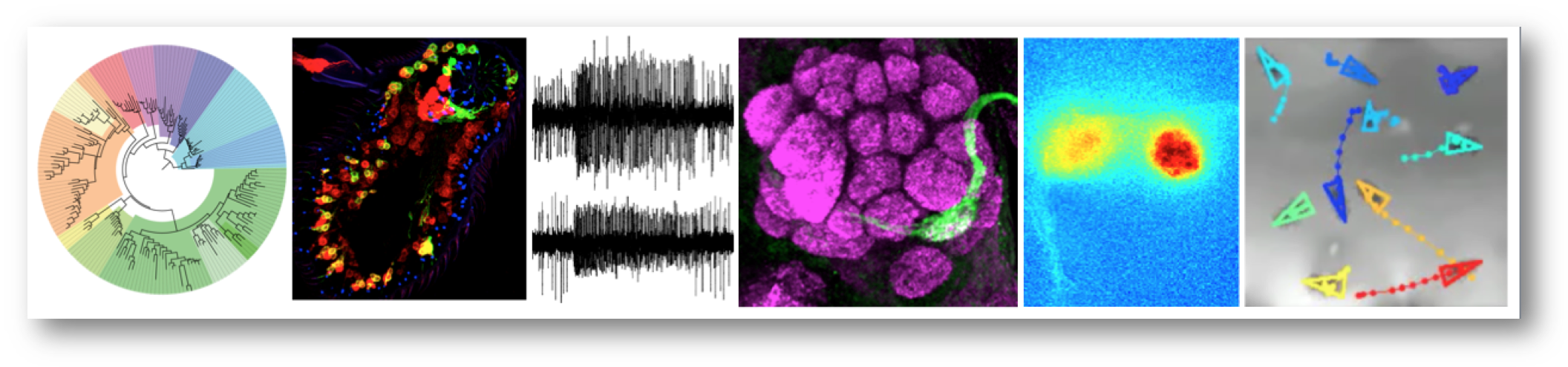
Research summary
Our group is interested in the structure, function and evolution of nervous systems. We focus on the olfactory system, which mediates recognition of myriad environmental signals to control diverse behaviours. As a model, we use Drosophila melanogaster, whose nervous system is sophisticated but numerically simple and experimentally highly tractable; many of our projects involve comparative functional approaches with both closely related drosophilid species and more evolutionarily distant invertebrates. We take a multidisciplinary approach, including bioinformatics, genetics, molecular and cellular biology, electrophysiology, optical imaging, and behavioural analysis. Our work provides fundamental insights into how nervous systems develop, operate and change, as well as offering opportunities to devise novel chemical sensors and rational strategies to control the odour-guided behaviours of insect vectors of disease and agricultural pests.
Representative publications
Shahandeh MP, Abuin L, Lescuyer De Decker L, Cergneux J, Koch R, Nagoshi E and Benton R. Circadian plasticity evolves via regulatory changes in a neuropeptide gene. Nature (2024) doi: 10.1038/s41586-024-08056-x
Takagi S, Sancer G, Abuin L, Stupski SD, Arguello JR, Prieto-Godino LL, Stern DL, Cruchet S, Álvarez-Ocaña R, Wienecke CFR, van Breugel F, Jeanne JM, Auer TO and Benton R. Olfactory sensory neuron population expansions influence projection neuron adaptation and enhance odour tracking. Nature Communications (2024) 15:7041Mika K, Cruchet S, Chai PC, Prieto-Godino LL, Auer TO, Pradervand S and Benton R. Olfactory receptor-dependent receptor repression in Drosophila. Science Advances (2021) 7(32):eabe3745
Our current work comprises three, interrelated research themes, outlined below. More information can be found in our publications, our pre-prints or by contacting us.
Structure, function and evolution of chemosensory receptors
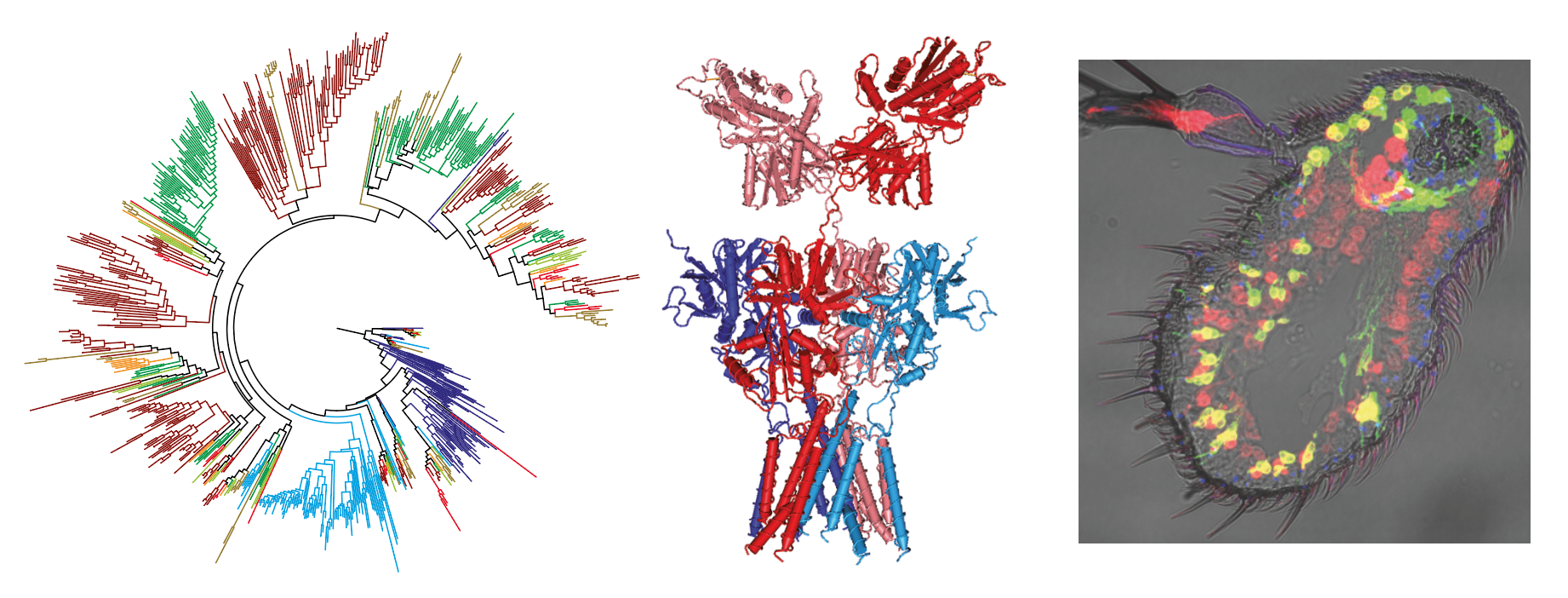
Since our discovery in 2009 of Ionotropic Receptors (IRs) – a subclade of the ionotropic glutamate receptor superfamily of ligand-gated ion channels – as a novel family of insect olfactory receptors, we and others have shown that these proteins have evolved diverse roles in olfaction, gustation, hygrosensation and thermosensation across insects. IRs form heteromeric complexes of “tuning” receptors, which define stimulus specificity, and one or more co-receptor subunits, which function in subcellular trafficking and signalling. We are currently developing in vivo, in vitro and in silico approaches to understand the mechanisms and evolution of ligand-recognition, heteromeric complex assembly and channel gating. We are also studying the evolutionary origin and functional diversification of a second family of insect chemosensory receptors, encompassing Odorant Receptors (ORs) and Gustatory Receptors (GRs).
Neurodevelopment and evolutionary plasticity of olfactory circuits
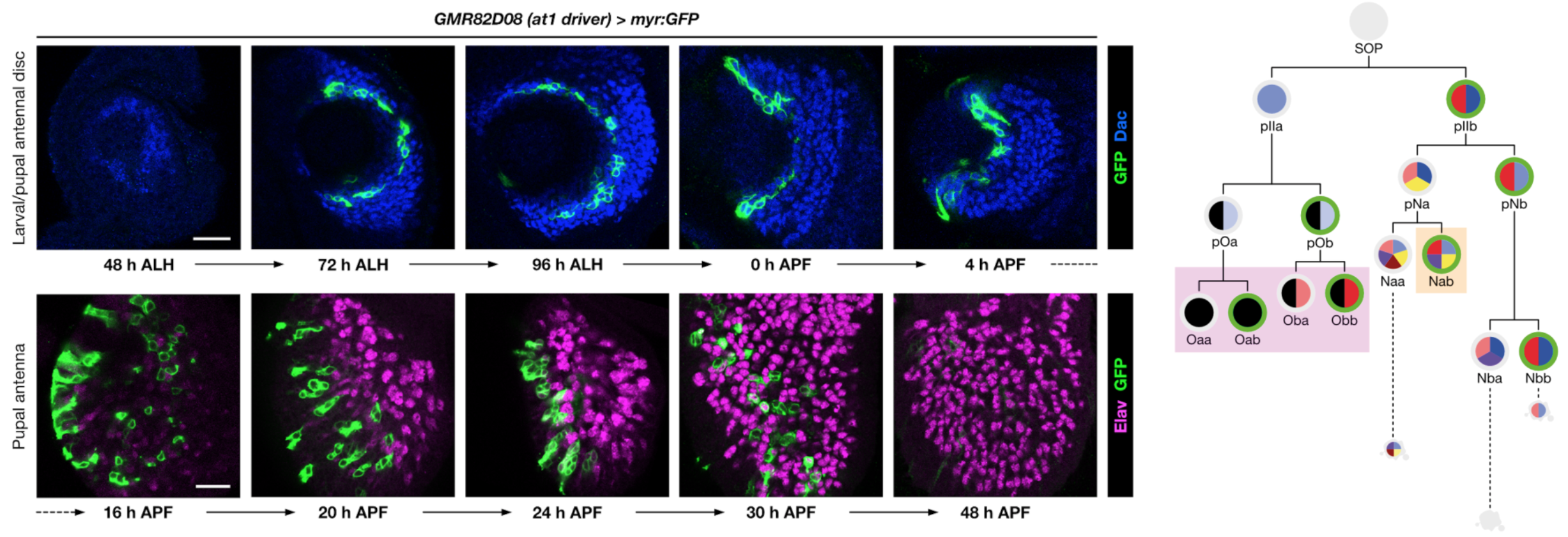
Olfactory pathways are one of the most dynamically evolving parts of the nervous system: animals frequently acquire (and discard) olfactory receptors, circuits and odour-evoked behaviours with the ever-changing landscape of stimuli in their environment. The evolutionary flexibility of olfactory systems is reflected in their modular organisation: in insects (as in vertebrates), most individual olfactory sensory neurons (OSNs) express just one olfactory receptor gene, and the axons of OSNs expressing the same receptor converge on discrete regions of neuropil (glomeruli) within the primary olfactory centre, where they synapse with second-order neurons. The numbers of olfactory receptors vary widely across species, with concordant diversity in the number and organisation of OSNs and glomeruli in the brain.
Using molecular genetic, single-cell sequencing and circuit tracing approaches in the D. melanogaster peripheral olfactory system, we are studying the mechanisms and evolution of (i) neuronal lineage specification, (ii) olfactory receptors’ singular expression patterns, (iii) how novel populations of OSNs arise through changes in patterns of neurogenesis and developmental programmed cell death, and (iv) how OSN populations are segregated to distinct glomeruli to form unique sensory channels in the brain. The mechanisms and molecules we characterise are likely to be relevant for understanding circuit formation and evolution in other brain regions and species.
Drosophila sechellia: a novel genetic model for behavioural evolution and neuro-ecology
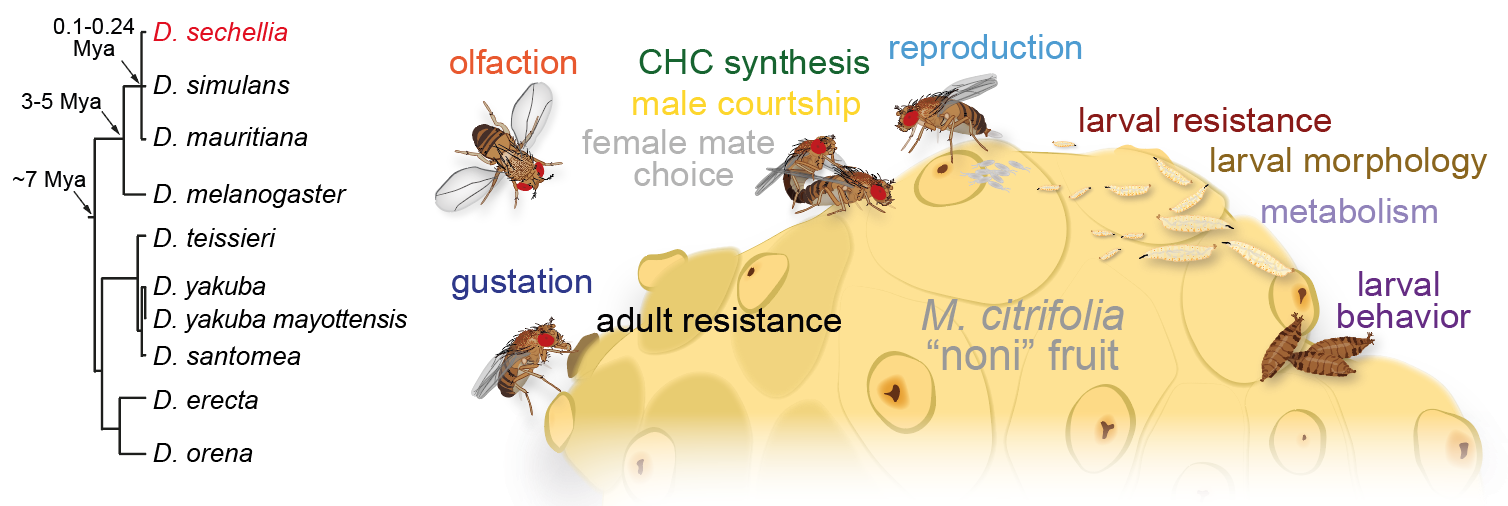
How animals’ extraordinarily diverse behaviours have evolved is unknown. Relating interspecific behavioural differences to anatomical or physiological distinctions in neural circuits, and causal genetic variation, offers a powerful approach to inform how nervous systems develop, function and change. We are establishing a new model neurogenetic system, Drosophila sechellia, an island endemic that is closely related to D. melanogaster and D. simulans. While D. sechellia retains global genomic and superficial morphological similarity to its cosmopolitan generalist cousins, this species has adapted to a unique ecological niche, using Morinda citrifolia "noni" fruit as a sole host for feeding and breeding. This work, which now takes our lab beyond the olfactory system, is being developed through three main aims:
(i) Establishment of a D. sechellia (neuro)genetic toolkit: we are building essential genetic reagents for generation and maintenance of animals of desired genotypes, for neurogenetic manipulations, and for recombination mapping-based approaches.
ii) Behavioural, neuroanatomical and molecular phenomics: systematic comparison of D. sechellia, D. simulans and D. melanogaster for their behaviours, their neuroanatomy and their neuro- molecular expression properties should reveal how D. sechellia has adapted to its niche, and will provide multiple entry-points to relate molecular, neuronal and behavioural differences between these species.
iii) Defining the genetic basis and functional significance of neuronal adaptations in D. sechellia: through high-resolution quantitative trait mapping and interspecific allele swap approaches, we aim to identify the causal genetic changes underlying neural adaptations in D. sechellia, and their physiological and behavioural significance.

Alumni: Benton lab members go on to diverse roles in science: independent research positions in academia or further research training, positions in industry, start-ups, science policy and education. Read more about what former lab members are up to here!
 |
Liliane Abuin - Technician Liliane obtained her Diplôme de Technicienne en Analyses Biomédicales from the Ecole Cantonale Vaudoise de Laborantins et Laborantines Médicaux. During 1996-2007 she worked in the group of Susanna Cotecchia in the Department of Pharmacology and Toxicology at the University of Lausanne. She has worked in the lab since October 2007. |
 |
Steeve Cruchet - Technician Steeve obtained his Diploma of Technician in Biomedical Analysis from the Ecole Cantonale Vaudoise de Laborantins et Laborantines Médicaux. From 2007 to 2010 he worked in the group of Thierry Pedrazzini in the Unity of Experimental Cardiology at the University Hospital of Lausanne. He joined the lab in March 2011. |
 |
Caroline Fragnière - Master Student Caroline attended high school near Lausanne, where she discovered biology, before travelling and working outside academia. She completed her Bachelors in Biology from the University of Lausanne and worked in our lab during summer 2023, supervised by Michele Marconcini, before pursuing her interests in neuroscience through a Master's degree at UNIL. She returned to our lab in Feb 2024 to pursue her Master research project. |
 |
Nathaniel Himmel - Post-doctoral Fellow Nate obtained his BS in biology from the University of Florida, and thereafter studied ion and water homeostasis at Emory University under Dr. Mitsi Blount. Nate’s PhD research at Georgia State University - in the lab of Dr. Dan Cox - concerned Drosophila cold and chemical sensing, insect cold acclimation, and the evolution of ion channels. Nate joined the lab in February 2022, funded by an HFSP Long-term Fellowship, to study the function of putative ligand-gated ion channels in unicellular eukaryotes, in the hopes of making inferences about the very early evolution and diversification of ion channels. Learn more about Nate on his website. |
 |
Michele Marconcini - Post-doctoral Fellow Michele obtained his Master's degree in molecular biology and genetics and his PhD in genetics, molecular and cellular biology at the University of Pavia. There, he first worked in the laboratory of Prof. Anna Malacrida, where he studied the population genetics of the invasive mosquito species Aedes albopictus, connected to the risk of arboviral disease outbreaks. Then, he worked in the Bonizzoni Lab, where he studied the evolution and the role of different sRNA pathways in the establishment of persistent viral infection in Aedes spp. mosquitoes. He joined our lab in October 2020 to initiate experimental evolution experiments in drosophilids. |
 |
Ambra Masuzzo - Post-doctoral Fellow Ambra obtained her PhD in the Royet lab at the University of Aix-Marseille, where she studied the impact of bacteria on Drosophila melanogaster's behaviors, including dissecting the cellular and molecular basis of bacteria-mediated oviposition suppression and gustatory detection of environmental bacteria. She joined our lab in August 2021 to examine inter-specific interactions underlying host specialization of Drosophila sechellia on Morinda citrifolia. She was awarded an EMBO Long-term Fellowship in June 2022. |
 |
Jérôme Mermet - Post-doctoral Fellow Jérôme obtained his Bachelor and Master degrees from University of Toulouse in France where he studied epigenetic regulation of gene expression in breast cancer cells in the lab of Kerstin Bystricky. He then joined the lab of Félix Naef at the EPFL, Switzerland, to study the dynamics of the three-dimensional organisation of chromatin across the circadian cycle and its function for circadian-clock gene transcription in mouse tissues. He joined our lab in April 2019 to investigate molecular mechanisms modulating olfactory circuit architecture during development in Drosophila. |
 |
Divyansh Mittal - Post-doctoral Fellow Divyansh obtained his Bachelors in Biomedical Science form Acharya Narendra Dev College, University of Delhi in 2015. He then joined the Indian Institute of Science (IISc), Bangalore for the Integrated Ph.D. program (Master and Ph.D. combined) in Biological Science. For his Ph.D., he worked under the guidance of Prof. Rishikesh Narayanan at the Molecular Biophysics Unit, IISc, and elucidated different neuronal- and network-scale mechanisms which impart functional robustness to distinct neural systems in the presence of heterogeneity and stochasticity. He joined our lab in November 2022. Divyansh is also part of the Early-Career Advisory Group at eLife. |
 |
Venkatesh Pal Mahadevan - Post-doctoral Fellow Venkatesh obtained his integrated Master’s degrees (M.Sc-M.Tech) in biotechnology from the University of Pune in 2016. During his Master’s program, he conducted his research project at the National Chemical Laboratory (NCL), focusing on the behavior of Aedes aegypti. He completed his PhD at the Max Planck Institute for Chemical Ecology in Jena (2024) under the supervision of Prof. Bill Hansson, where he investigated the neuroecology of two non-model drosophilids (D. virilis and D. busckii). Venkatesh will join our lab in January 2025 to study the evolution of neural circuits and tolerance mechanisms to noxious chemicals in drosophilids. |
 |
Asfa Sabrin Borbora - PhD Student Asfa obtained her Master's degree from the Tata Institute of Fundamental Research in India, where she worked with Prof. Sreelaja Nair. There, she worked on how cell sizes evolve in early zebrafish development and how sizes of dynamic cell organelles such as the mitotic spindle and centrosome scale during reductive cell divisions. She joined our lab in December 2021, where she will work at the interface of developmental biology and neuroscience to understand neuronal connection specificity in the Drosophila olfactory system. |
 |
Suguru Takagi - Post-doctoral Fellow Suguru obtained his BEng from the Department of Applied Physics, University of Tokyo, where he studied the properties of shot noise in interferometry using power modulated laser beam. He then obtained his MS and PhD from the Department of Physics, the University of Tokyo, where he studied the circuit mechanisms and their molecular underpinnings of action selection in Drosophila melanogaster larvae, in collaboration with investigators in HHMI Janelia Research Campus and RIKEN BSI. He joined the lab in September 2019 to carry out cross-species comparisons of central olfactory circuits in drosophilids, supported by an EMBO Long-Term Fellowship and a Marie Sklodowska-Curie Individual Fellowship. Learn more about Suguru here. |
 |
Giovanna Zappia - Technician Giovanna obtained her MS in Pharmaceutical Biotechnology from the University of Milan, Italy, where she studied the role of ADAM10, a metalloprotease involved in the physiological functioning, brain development and pathogenesis of Alzheimer's disease. In 2010 she joined the Andrea Volterra's Lab in the Department of Fundamental Neuroscience at UNIL where she investigated the role of TNFα on the astrocytes in pathological conditions, particularly in experimental autoimmune encephalomyelitis (EAE), a mouse model of Multiple Sclerosis. She joined our group in Oct 2013. |
The Benton lab is active in developing, optimising and documenting a number of diverse experimental methods we (and others) use. Please see below for links to published articles.
Bioinformatics
We have collaborated with Debbie Marks' lab at Harvard to generate the first de novo three-dimensional models of insect Ors using EVfold-Transmembrane. You can find all the resources for this method on the EVfold webpage.
Reference: Hopf TA, Morinaga S, Ihara S, Touhara K, Marks DS and Benton R. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nature Communications (2015) 6:6077
Our more recent modelling approaches, have used de novo protein folding algorithms to identity potential homology between insect Ors, and polytopic membrane proteins from unicellular eukaryotes and plant DUF3537 proteins as described in our paper: Benton R, Dessimoz C, Moi D. A putative origin of the insect chemosensory receptor superfamily in the last common eukaryotic ancestor. eLife (2020) 9:e62507
Full outputs of the modelling analyses are available on the Dryad repository
Histology and imaging
Saina M and Benton R. Visualizing olfactory receptor expression and localization in Drosophila. In Methods in Molecular Biology (2013) Vol. 1003
Greg Jefferis' lab at the MRC-LMB has developed powerful tools for chemical-tag based in situ labelling; we've shown that these work well also in peripheral appendages:
Sutcliffe B, Ng J, Auer TO, Pasche M, Benton R, Jefferis GSXE and Cachero S. Second Generation Chemical Tags: Sensitivity, Versatility and Speed. Genetics (2017) doi:10.1534/genetics.116.199281
Electrophysiology
Benton R and Dahanukar A. Chemosensory coding in single sensilla. Cold Spring Harbor Protocols (2022) doi:10.1101/pdb.top107803
Benton R and Dahanukar A. Recording from fly olfactory sensilla. Cold Spring Harbor Protocols (2022) doi:10.1101/pdb.prot108063
Dahanukar A and Benton R. Recording from fly taste sensilla. Cold Spring Harbor Protocols (2022) doi: 10.1101/pdb.prot108064
Optical imaging
Silbering AF, Bell R, Galizia CG and Benton R. Calcium imaging in the Drosophila antennal lobe. Journal of Visualized Experiments (2012) 10.3791/2976
Behaviour
Ramdya P, Schaffter T, Floreano D and Benton R. Fluorescence Behavioral Imaging (FBI) tracks identity in heterogeneous groups of Drosophila. PLOS ONE (2012) 7(11):e48381
See also this resource page for FBI
Uhlmann V, Ramdya P, Delgado-Gonzalo R, Benton R and Unser M. FlyLimbTracker: an active contour based approach for leg segment tracking in unmarked, freely behaving Drosophila. PLOS ONE (2017) 12(4):e0173433
Blueprints for the single-fly oviposition assay based upon the design from the Yang lab, used in Álvarez-Ocaña R, Shahandeh MP, Ray V, Auer TO, Gompel N, and Benton R. Odor-regulated oviposition behavior in an ecological specialist. Nature Communications (2023) 14:e3041.
Genetic tools in non-melanogaster drosophilids
We are currently generating many genetic & neurogenetic tools in Drosophila sechellia (as well as a few in other drosophilid species) as part of our comparative neuroscience projects. See our recent paper for more details on some of these efforts:
Olfactory receptor and circuit evolution promote host specialization. Nature (2020) 579, 402-408
Reference brains for D. sechellia females and males, generated by Richard Benton and Greg Jefferis are available here
Tools in other species:
D. simulans: Auer TO, Álvarez-Ocaña R, Cruchet S, Benton R, Arguello JR. Copy number changes in co-expressed odorant receptor genes enables selection for sensory differences in drosophilid species. Nature Ecology and Evolution (2022) 6, 1343–1353
D. mojavensis: Khallaf MA, Auer TO, Grabe V, Depetris-Chauvin A, Ammagarahalli B, Zhang D-D, Lavista-Llanos S, Kaftan F, Weißflog J, Matzkin LM, Rollmann SM, Löfstedt C, Svatos A, Dweck HKM, Sachse S, Benton R, Hansson BS and Knaden M. Mate discrimination among subspecies through a conserved olfactory pathway. Science Advances (2020) 6(25):eaba5279
We are interested in establishing community exchange for sharing ideas and reagents in genetic modification of non-melanogaster insect species. Please contact Richard Benton for more information.
Open Labware / 3D Printing
Through her work with TReND in Africa, Lucia Prieto-Godino has been involved in development of 3D design and printing for labs:
Baden T, Chagas AM, Gage G, Marzullo T, Prieto-Godino LL, Euler T. PLOS Biol (2015) 13(3):e1002086
Maia Chagas A, Prieto-Godino L, Arrenberg AB and Baden T. The €100 lab: A 3D-printable open-source platform for fluorescence microscopy, optogenetics, and accurate temperature control during behaviour of zebrafish, Drosophila, and Caenorhabditis elegans. PLOS Biol (2017) 15(7):e2002702
Some of the 3D printed object designs we use in the research in our group are available through Thingiverse
Datasets
Bulk RNA-Seq of control and pointedRNAi antennae (GSE113997) from: Sensory neuron lineage mapping and manipulation in the Drosophila olfactory system". Chai P.C., Cruchet S., Wigger L., Benton R. Nature Communications (2019) 10 (1) p. 643
Bulk RNA-Seq of control and peb-Gal4>UAS-p35 antennae (GSE128725) from: Prieto-Godino LL, Silbering AF, Khallaf MA, Cruchet S, Bojkowska K, Pradervand S, Hansson BS, Knaden M and Benton R. Functional integration of “undead” neurons in the olfactory system. Science Advances (2020) 6(11):eaaz7238
TaDa sequencing data from seven Ir OSN populations in ArrayExpress database at EMBL-EBI (E-MTAB-8935); the workflow and corresponding code is available on GitLab from: Arguello JR, Abuin L, Armida J, Mika K, Chai PC, Benton R. Targeted molecular profiling of rare olfactory sensory neurons identifies fate, wiring and functional determinants. eLife (2021) 10:e63036
Bulk RNA-Seq of control and E93RNAi antennae (GSE150296) from: Mika K, Cruchet S, Chai PC, Prieto-Godino LL, Auer TO, Pradervand S and Benton R. Olfactory receptor-dependent receptor repression in Drosophila. Science Advances (2021) 7(32):eabe3745
Two comparative transcriptomic datasets listed below are described in: Scalzotto M, Ng R, Cruchet S, Saina M, Armida J, Su C-Y, Benton R. Pheromone sensing in Drosophila requires support cell-expressed Osiris 8. BMC Biology (2022) 20(1):230
- Comparative transcriptomic analysis of the D. melanogaster antennal olfactory subsystems (GSE183763), Saina M and Benton R. We identified genes whose expression is enriched in the D. melanogaster antennal olfactory subsystems that express Odorant receptors or Ionotropic receptors by microarray analysis of RNA extracted from antennae of wild-type (Oregon R P2) animals, ato mutants (which lack the Ir subsystem) and amos mutants (which lack the Or subsystem).
- Transcriptomic analysis of wild-type and amos mutant D. melanogaster third-instar larval antennal discs and adult antennae (GSE190696), Armida J and Benton R.
Quelques articles/emissions sur notre recherche - Some articles/broadcasts about our research
Understanding the fruit fly's nose (SNF/Latsis) (en anglais, sous-titré en français)
Une mouche alliée de la science (RTS - CQFD)
What fruit flies can teach us about the science of smell (Podcast at swissinfo.ch)
Sa majesté des mouches (Le Temps)
Rencontre avec Richard Benton (RTS - CQFD)
Im Kopf von Fruchtfliegen - Migros Magazin (auf deutsch)
The wisdom of the fly crowds (Ed Yong/National Geographic)
The world's first true aphrodisiac (NBC News)
Our European Research Councial project on the evolution of olfactory circuits (english / français)
Pseudo-pseudogenes (or "zombie genes") - RTS1 video report, Le Matin, 24heures (en français); Nature Podcast (in english)
An ant’s kiss may hide a sneaky form of communication - a comment in Science on our paper on trophallaxis and chemical communication in social insects; see also the Daily Mail and Wire
Chacun son cerveau : l'évolution de la perception sensorielle - conférence du Prof. Richard Benton
Les mouches - RTS1 Brouhaha (émission pour enfants)

