Béatrice Desvergne was trained as a MD. She initially specialized in Anesthesiology and Resuscitation, practiced medicine for a few years, and decided to move for fundamental research. She then carried out a post-doctoral stay from 1988 to 1992 at the National Institutes of Health in Bethesda, first as visiting fellow and then visiting associate in the National Institute of Diabetes and Digestive and Kidney Diseases. In 1992, she was appointed as Assistant Professor at the Institute of Animal Biology of the UNIL. After being appointed as Associate Professor, she was promoted as full Professor in 2008. She joined the Center for Integrative Genomics in 2003.
Peroxisome proliferator-activated receptor, development, tissue repair, energy homeostasis, signal transduction
Research summary
Networking activity of PPARs during development and in adult metabolic homeostasis
As they mediate intracellular hormone action, nuclear receptors play a crucial multi-faceted role in coordinating growth during development, and homeostasis at adult stage. Among them, the peroxisome-proliferator activated receptors (PPARs) act as fatty acids sensors, responding to dietary as well as to endogenous challenges. Accordingly, they have an integrative role in controlling the expression of genes regulating the storage, mobilization, and/or utilization of lipids. Using various molecular, cellular, and animal approaches, our studies are aimed at understanding how PPARs are integrated in the main pathways that shape the organism during development on the one hand and maintain systemic homeostasis on the other hand.
We were among the first to generate PPAR mutant mice. Following a clinician-type of approach, our activities have been centered on revealing and understanding at the molecular levels the phenotypic expressions of PPAR mutations, taking them as leads to explore the physiopathological significance and novel therapeutic advances that PPARs carry.
Our studies during development show that both PPARβ and PPARγ are required for placenta development but each has specific activities: on the differentiation of trophoblast giant cells, via activation of the PI3K pathway and inhibition of Id2 for PPARβ, and on vascular development via controlling the expression of angiogenic factors for PPARγ. Further studies of the role of PPARβ and PPARγ in development were impeded by a partially and fully penetrant lethality of PPARβ-/- and PPARγ-/- embryos, respectively, at embryonic day 10.5. However, we have now generated fully viable mutant embryos and live pups, through an epiblastic selective PPAR deletion. It demonstrates that the cause of embryonic lethality in PPAR mutants is mainly due to the placental defects and gives us new tols for exploring the role of PPARγ in late development and in adult tissues.
In the adult animals, the gut is a very interesting organ, combining critical metabolic functions and a tightly regulated and highly active cell renewal capacity, from few adult stem cells to highly differentiated enterocytes, Paneth, Goblet, and enterodendocrine cells. In this tissue, we demonstrated that decrease of Indian Hedgehog via PPARβ ensures the final maturation of Paneth cell precursors whereas PPARγ is rather involved in controlling inflammation processes. We are now further exploring the importance of PPARs in response to challenges, either metabolic, infectious, or physical damages, with the purpose of bringing our mechanistic approaches in mouse models closer to human pathologies.
The role of PPARs as mediating the activity of some endocrine disruptors was an important focus of our recent activity. To understand how endocrine disruptors behave at the molecular levels in the living cells, we used a combination of Fluorescence Recovery After Photobleaching (FRAP), Fluorescence Correlation Spectroscopy (FCS) and Fluorescence Resonance Energy Transfer (FRET). We first demonstrated that PPARs readily heterodimerize with retinoid X receptor (RXR) and exhibit a ligand-induced reduction of mobility, probably due to enhanced interactions with cofactors and/or chromatin. We also demonstrated that coregulator recruitment (and not DNA binding) plays a crucial role in receptor mobility, suggesting that transcriptional complexes are formed prior to promoter binding. This allowed us to demonstrate that, in the living cells, the pollutant monoethylhexyl-
phthalate (MEHP) directly binds to the ligand binding domain of PPAR and drives the recruitement of a specific subset of cofactors. We further analyzed the consequences in vivo of such activities and showed that MEHP protects mice from diet-induced obesity via a PPARα-dependent activation of hepatic fatty acid catabolism. This is accompanied by, and possibly due to, the up-regulation of the anti-obesity factor FGF21 hepatic expression. However, both effects are reversed in PPARα-humanized mice, underlining the importance of PPARα species-specific activities and questioning the impact of phthalates in human metabolic homeostasis.
Representative publications
F. Varnat, B. Bordier-ten Heggeler, P. Grisel, N. Boucard, I. Corthésy-Theulaz, W. Wahli, and B. Desvergne. (2006) PPARbeta/delta regulates Paneth cell differentiation via controlling the hedgehog signaling pathway. Gastroentrology 131:538-53
Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. (2006) Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor beta/delta. Mol Cell Biol. 26:3266-81.
B. Desvergne, L. Michalik, W. Wahli. (2006) Transcriptional control of metabolism. Physiological Review, 86:465-514.
Feige JN, Gelman L, Tudor C, Engelborghs Y, Wahli W, Desvergne B. (2005). Fluorescence imaging reveals the nuclear behavior of PPAR/RXR heterodimers in the absence and presence of ligand. J Biol Chem. 280:17880-90.
E. Letavernier, J. Perez, E. Joye, A. Bellocq, B. Fouqueray, J-P Haymann, D. Heudes, W. Wahli, B. Desvergne, and L. Baud. (2005). PPARbeta/delta exerts a strong protection from ischemic acute renal failure. J Am Soc Nephrol. 16:2395-402.
Advanced search is available through Serval
Publications can be managed by accessing Serval via MyUnil
Publications
PixFRET, an ImageJ plug-in for FRET calculation which can accommodate variations in spectral bleed-throughs
Jérôme Feige1, Daniel Sage2, Walter Wahli1, Béatrice Desvergne1 and Laurent Gelman1
1 - Center for Integrative Genomics, NCCR frontiers in Genetics, University of Lausanne, Switzerland.
2 - Biomedical Imaging Group (BIG), Swiss Federal Institute of Technology Lausanne (EPFL), Lausanne, Switzerland.
Fluorescence Resonance Energy Transfer (FRET) is a technique used to investigate interactions between fluorescent partners that has gained great interest for cell biologists with the recent introduction of auto-fluorescent proteins which can be coupled to a protein of interest to produce a fluorescent chimera.
A plethora of methods exists to calculate FRET, depending on the protocol used (e.g. sensitized emission versus acceptor photobleaching) and the precision that is pursued (for a review, see "FRET or no FRET: a quantitative comparison", Biophysical J., 2003:84 p.3992-4010).
The ImageJ plug-in PixFRET allows to generate images of FRET, and hence to visualize FRETwithin a cell or a cell population, by computing pixel by pixel the images of a sample acquired in a three channel setting, according to the formula and the methodology described by Gordon et al (Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy, Biophysical J., 1998:74(5), p.2702-13) and Xia and Liu ("Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes", Biophysical J., 2001:81(4), p.2395-402).
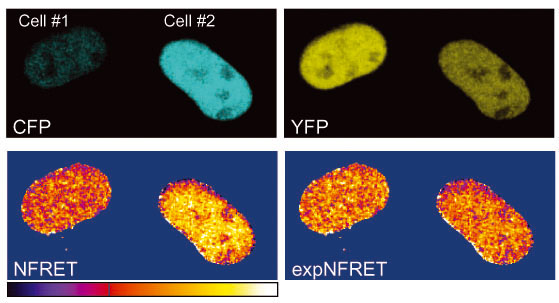
ExpNFRET reduces inter-cellular variability in pixel-by-pixel analyses.
Plug-in installation
- Download the PixFRET.zip file
- Double click on the PixFRET.zip icon to extract the plug-in. This will create a folder with two new files. The ".jar" file is the plug-in and the ".pdf" file the user's guide.
- Place the ".jar" file in the "Plugins" folder in ImageJ.
- Start ImageJ again to display the new plug-in in the "Plugins" menu.
Note that ImageJ is a public-domain software package for image processing. It is free and it doesn't take more than a couple of minutes to install (http.//rsb.info.nih.gov/ij), it runs on any platforms: Unix, Linux, Windows, Mac OS9, Mac OSX.
PixFRET2006-05-17.zip (357 Ko)


