Bassani-Sternberg Lab
Our focus
Our main goal is to identify clinically relevant cancer specific Human Leukocyte Antigen (HLA) ligands that will guide the development of personalized cancer immunotherapy using state of the art proteogenomics and mass-spectrometry (MS) approaches.
Our projects
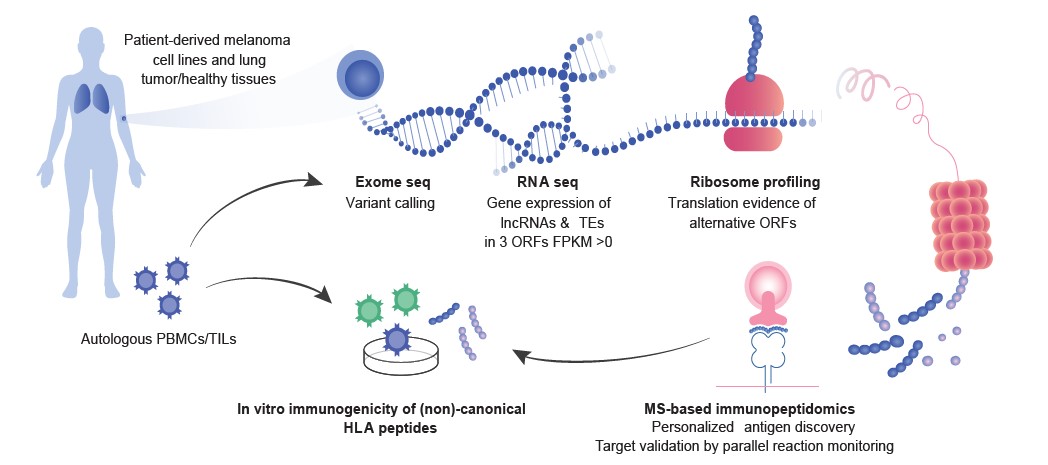
- We develop proteogenomics and MS-based immunopeptidomics analytical and computational approaches to identify HLA ligands derived from tumor-associated proteins, mutated neoantigens, non-canonical ORFs and post translationally modified peptides.
- We have initiated fundamental discovery work to elucidate how tumor cells present antigens and what are the bases of tumor immunogenicity in mouse and human.
- We are investigating the differences between T cell inflamed and non-inflamed tumors in terms of antigen presentation and the antigenic landscape and how drugs modulate the immunopeptidome.
- We have established a continuous bioinformatics pipeline called NeoDisc enabling direct and personalized identification and prediction of neoantigens by combining genomic information derived from exome-seq analysis and transcriptomics data with measured immunopeptidomics data.
- We apply our discovery pipeline in the context of phase I clinical vaccine and adoptive T cell therapy trials at the CHUV to identify personalized neoantigens from patient tumor samples.

- Müller M.*, Huber F., Arnaud M., Kraemer A., Ricart Altimiras E., Michaux J., Taillandier-Coindard M., Chiffelle J., Murgues B., Gehret T., Auger A., Stevenson B.J., Coukos G., Harari A., Bassani-Sternberg M* Machine learning methods and harmonized datasets improve immunogenic neoantigen prediction. Immunity *Co-corresponding authors (2023)
- Kraemer A., Chong C., Huber F., Pak H., Stevenson B.J., Muller M., Michaux J, Ricart Altimiras E., Rusakiewicz S., Simó-Riudalbas L., Planet E., Wiznerowicz M., Dagher J., Trono D., Coukos G., Tissot S., Bassani-Sternberg M* The immunopeptidome landscape associated with T cell infiltration, inflammation and immune-editing in lung cancer. Nature Cancer *Corresponding author (2023)
- Chong C., Coukos G., Bassani-Sternberg M* Identification of tumor antigens with immunopeptidomics. Nature Biotechnology *Corresponding author (2022)
- Pak, H., J. Michaux, F. Huber, C. Chong, B. J. Stevenson, M. Muller, G. Coukos and M. Bassani-Sternberg* Sensitive Immunopeptidomics by Leveraging Available Large-Scale Multi-HLA Spectral Libraries, Data-Independent Acquisition, and MS/MS Prediction. Molecular & Cellular Proteomics *Corresponding author (2021)
- , , , , , , , , , , , , , , , , , , , , , , , , , Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nature Communications *Corresponding author (2020)
Meet all the Bassani Group members.
|
Funding |
|
|
Affiliations |
|
|
Links |
|







.png)


