DELORENZI Lab
- : -
Mauro Delorenzi retired in March 2022.
- : -
Focus of Delorenzi Lab
- Supervision and collaboration for bioinformatics activities in oncology research
- Discovery-oriented collaborations in fundamental and clinical cancer research
- Development and validation of prognostic and predictive signatures
- Development and comparative testing of methods
- Training in statistics & statistical computing with applications (elementary to advanced)
Projects of Delorenzi Lab
Methods for data exploration and visualization
One approach focuses on improving computational methods that are important to more efficiently investigate and visualize relationships among data, across variables describing samples (like mutations or gene expression) and outcomes of interest (like patient survival or response to treatment). The aim is to allow the analyst to quickly extract and understand the most salient information in the data, at the level of interesting explanatory power and appropriate mathematical models, as well as at the level of potential quality issues and of strategies of data adjustments or filtering and stratification.
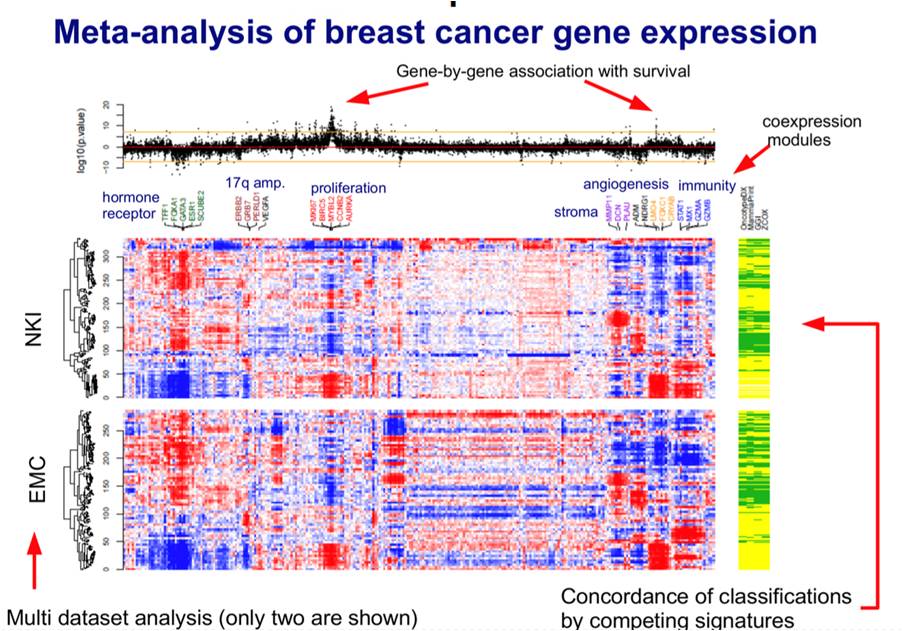
Investigation of consistent relations between gene-expression data and survival signatures over multiple independent datasets.
Signatures: Methods for Translating from Research to Application
We think that a main reason of difficulty in translating multi-variable prediction models from research to clinical settings is projects typically rely on intricate and unstable processing steps to provide supposedly bias-free datasets to discover and train new omics signatures.
But in unprocessed data, some variables likely capture systematic biases. These relations can be exploited in a predictor to render it more robust to changing measurement conditions. We develop statistical approaches suitable for practical applications of this principle of “self-normalizing predictors”, which we believe might turn out to be more easily translated to applications, especially if clinical applications are aimed at, where it is crucial to be able to reliably analyze single samples one by one in real time as they enter the clinic.
Molecular characterization of cancer subtypes and exploration of treatment efficacy signatures
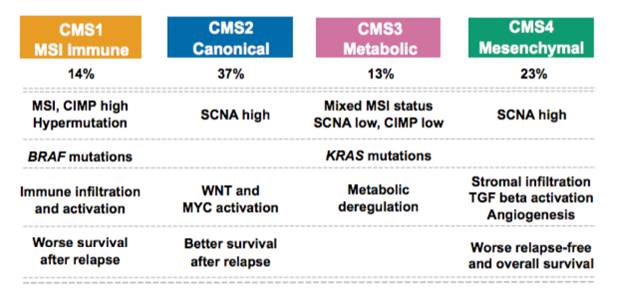
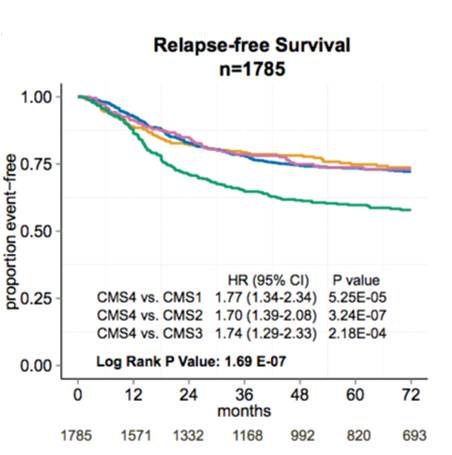
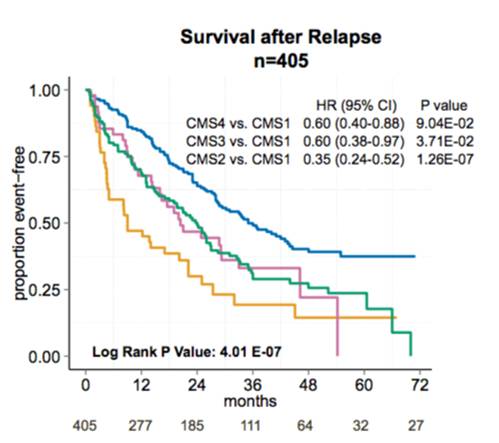
We have explored heterogeneity of gene expression in various cancer types, in particular primary colon cancer, summarized it with a system of five subtypes (of which one formed by tumors with a mixed profile) and later contributed to an international consortium that established a consensus of four well characterized molecular subtypes CMS1-CMS4 and a group of unclassified not well characterized tumors.
Ongoing efforts aim at expanding the study to metastatic colon cancer and to other cancer types.
We help clinical and pharma groups in the analysis of gene expression and genetic profiles of tumors, especially in the retrospective and exploratory investigation of clinical trials data, searching for significant relations between tumor features and treatment efficacies. The aim is to test if any biomarkers of response can be identified and then validated in successive studies.
 |
 |
KEY PUBLICATIONS

- Dangaj D, et al. Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors. 2019 Cancer Cell. 2019;35(6):885-900.e10. PMID: 30699351
- Mooi JK, Wirapati P, et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. 2018 Ann. Oncol. 2018;29(11):2240-2246. PMID: 3024752
- Cardoso F et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. 2016 N Engl J Med. 2016;25;375(8):717-29. PMID: 27557300




